Abstract
Purpose: Molecular genetic knowledge of clear-cell renal-cell carcinoma (CCRCC) plays an important role in predicting the prognosis and may be used as a guide in treatment decisions and the conception of clinical trials. It would then be desirable to predict these mutations non-invasively from CT images which are already available for CCRCC patients. Methods: TCGAKIRC data were obtained from the National Cancer Institute’s (NCI) image dataset. We used 191 patient data of which 63 were associated with PBRM1 mutations. The tumors were delineated by a radiologist with over 10 years of experience, on slices that displayed the largest diameter of the tumor. Features were extracted and normalized. After feature selection, the KNN classification with Random Subspace method was used as it is known to have advantages over the simple k-nearest-neighbor method. Results: Prediction accuracy for PBRM1 was found 83.8 %. Conclusions: A single slice of the CT scan image of CCRCC can be used for predicting PBRM1 mutations using KNN classification in Random Subspaces with an acceptable accuracy.
Highlights
- Polybromo-1 (PBRM1) mutation of a clear cell renal cell carcinoma can be predicted from CT images
- KNN classification in random subspaces gives fairly accurate results
- A relatively large number of patient images were used to improve reliability
1. Introduction
Renal Cell Carcinoma (RCC) is the most encountered type of renal cancer and represents about 3.7 % of new cancer occurrences. Just in The United States, RCC accounted for about 61.560 new patients and 14.080 deaths in year 2015. It is known that certain genes which have mutations can activate intracellular molecular pathways. These specific pathways lead to an increased risk of specific histological subtypes of RCC. This knowledge has helped us to better understand the pathogenesis of RCC and RCC has been divided into subtypes related to genetic structure and mutation status. According to the WHO, there are eight major subtypes of adult-onset RCC. Clear cell Renal Cell Carcinoma (CCRCC) is the most encountered one. Approximately 20 % of patients have metastatic disease at presentation. More than half of the patients develop metastases after the initial diagnosis.
Recently, the comprehension of the genetic base of RCC has improved research and led to the discovery of novel anticancer agents targeting specific intracellular pathways [1].
Polybromo-1 (PBRM1) gene is the second most common mutation and is seen in 40 % of these patients. The mutation encodes the protein BRG1-associated factor (BAF) 180 [2]. It affects the critical cellular processes by regulating cell-cycle changes, metabolism, and DNA repair [3].
There have been studies indicating that this gene is valuable because it has an impact on survival [4]. One of the studies about PBRM1 indicated that decrease of PBRM1 expression is linked with a bad prognosis and increased clinicopathological features in patients with RCC [5]. Another study with RCC stage-4 patients reported that this gene could have potential as a prognostic marker for advanced RCC [6]. Moreover, other studies indicate that the PBRM1 mutation status has a great potential to identify CCRCC and has noticeable effects on disease progression [7, 8] and may affect the new treatment strategies [9].
Recently several studies concentrated on gene mutation prediction for cancer patients from medical images noninvasively [10]. Few studies focused on the prediction of the PBRM1 gene mutation for CCRCC patients from imaging studies [11,12,13]. In [11], associations between imaging features determined by radiologists and the genetic mutation status were found. In [12], a multi-classifier multi-objective radiogenomics model was developed. In [13], an artificial neural network (ANN) and a random forest (RF) algorithm were used for classification. Since the original dataset was small, multiple slices were used for feature extraction in order to increase the sample size.
A general challenge in these studies is to deal with small and noisy datasets.
In this study, the goal was to address these issues and develop a method for predicting the PBRM1 gene mutation non-invasively using CT images and the KNN classification method with Random Subspaces for the first time. KNN is known to have several advantages such as simplicity and performance. Random Subspace Method for kNN Classification was shown to improve accuracy [14]. This superiority was demonstrated to be preserved with even smaller number of training samples, a condition often encountered with radiogenomic data.
The main novel contributions of this work are:
– The KNN method in Random Subspaces has been applied to CCRCC Radiogenomics for the first time,
– A relatively large number of patient data has been used (259). With small number of samples, the results may largely depend on the used dataset. Other studies have used relatively small overall number of patients (e.g. 45 in [13]).
In the following, the methods used for predicting the PBRM1 gene mutation will be outlined. First the data used will be described. Then, image processing steps will be given. Results from classification will be discussed in view of the literature and necessary future work will be indicated.
2. Methods
2.1. Data
The partnership between the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) has provided information on key genomic alterations in 33 identified cancer types, including CCRCC [15, 16]. TCGA-KIRC data set which has the disease type Adenomas and Adenocarcinomas of Kidney was used for this study. We used 191 scans from the dataset. 63 patients had the PBRM1 mutation.
2.2. Data processing
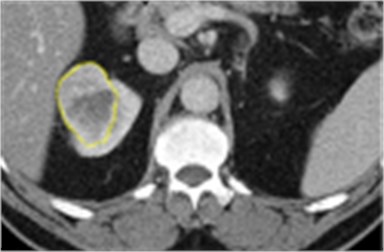
The tissue of interest was carefully delineated by a radiologist with over 10 years old experiences using ImageJ [17] software. The slices were considered to include the largest tumor area for each patient obtained from TCGA-KIRC data. A sample ROI is shown in Fig. 1. After the regions of interest (ROI) were drawn, radiogenomic features were extracted. These included gray level patterns, inter-voxel relationships, shape and texture features. In this step, 136 radiographic features were generated, and a high dimensional feature matrix was created. The software platforms Image J with texture analyzer plugin [18], MIPAV [19] and LifeX [20] were used.
Extracted radiogenomic features included shape, intensity and texture features. Unbalanced data were [21] handled for balancing by using the ADASYN algorithm which represents an extended version of SMOTE. After -tests on individual features, and neighborhood component analysis (FSCNCA) [22], the reduced number of features obtained was nine.
After the feature selection process, classifications were performed using Matlab R2019a. The confusion matrix was used to evaluate the results. Validation selection was set to 5-Fold cross validation (5). The selected model type was KNN with Random Subspace [14].
Fig. 1The tumor in the right kidney which had the PBRM1 mutation is indicated by the yellow contour
3. Results
Our results showed that using classification learner, KNN with Random Subspace model can correctly predict PBRM1 and NON-PBRM1 data with 83.8 % (see Figs. 2 and 3).
Fig. 2Confusion matrix for PBRM1 mutation status using KNN with random subspace
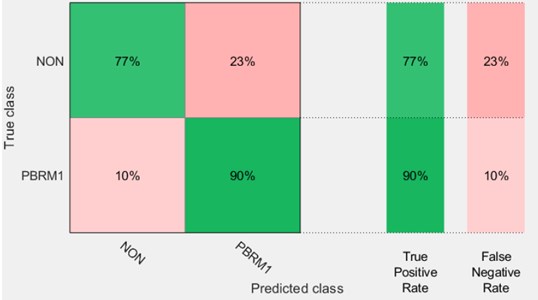
Fig. 3Number of observations for PBMR1 mutation status using KNN with random subspace
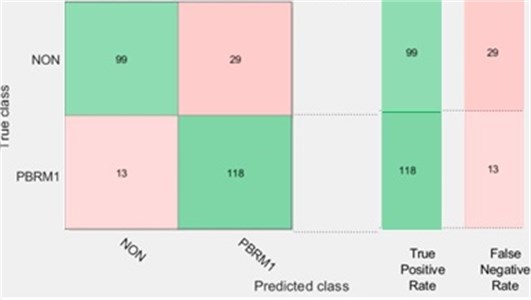
The results have been obtained using the Fine KNN model Random Subspaces in Matlab R2019a (1) using the Classification Learner, Distance metric = Euclidean, Distance weight = Equal, Subspace dimension = 6, Number of Learning Cycles = 30. Validation selection was set to 5-Fold cross validation (5).
4. Discussion
The goal of this study was to predict the PBRM1 gene mutation noninvasively using a single slice of a CT Image study for CCRCC patients. Predicting gene mutations is important for prognostic and therapy selection purposes. KNN classification with random subspace was used. The results of the study were presented in Fig. 2 and Fig. 3. These results show that the PBRM1 gene mutation could be predicted with a sensitivity of 90 % and a specificity of 77.5 %. Overall accuracy was determined as 83.8 %.
There is some evidence that these mutations influence the response to therapy [23]. Therefore, the implication of this study is that the non-invasive nature and the practical single slice approach of this technique may make it very useful in therapy selection.
The accuracy obtained is similar to the one obtained in a previous study (78 % in [12]). Up to 95 % accuracy was reported in another study [13] conducted with a total of 45 patients. However, the main disadvantage of these studies was the low number of patients and the lack of a large independent test dataset.
The study presented here was conducted with a larger patient dataset (191). However, the images were not standardized. Data included images from different CT scanners, contrast and non-contrast images.
In the future, using larger and more uniform datasets could increase the present performance. Practical implementation may require quick and accurate 3D methods for tumor delineation. Studies with better automatic 3-D segmentation algorithms may increase both the speed and the performance of these algorithms.
5. Conclusions
A practical machine learning implementation has been realized for the non-invasive determination of the mutation status of PBRM1 Mutation for Clear Cell Renal Cell Carcinoma patients. The KNN with Random Subspace was shown to be an adequate method for this task. More research is needed to increase both the accuracy and speed by using automated 3D segmentation algorithms and larger and standard datasets.
References
-
Shinagare A. B., Krajewski K. M., Braschi Amirfarzan M., Ramaiya N. H. Advanced renal cell carcinoma: role of the radiologist in the era of precision medicine. Radiology, Vol. 284, Issue 2, 2017, p. 333-351.
-
Le V. H., Hsieh J. J. Genomics and genetics of clear cell renal cell carcinoma: a mini-review. Journal of Translational Genetics and Genomics, Vol. 2, 2018, p. 17.
-
Nargund A. M., Osmanbeyoglu H. U., Cheng E. H., Hsieh J. J. SWI/SNF tumor suppressor gene PBRM1/BAF180 in human clear cell kidney cancer. Molecular and Cellular Oncology, Vol. 4, Issue 4, 2017, p. 1342747.
-
Kapur Payal, Samuel Peña Llopis, Alana Christie, Leah Zhrebker, Andrea Pavía Jiménez, Kimryn Rathmell W., Xian Jin Xie, Brugarolas James Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. The Lancet Oncology, Vol. 14, Issue 2, 2013, p. 159-167.
-
Wang Z., Peng S., Guo L., Xie H., Wang A., Shang Z., Niu Y. Prognostic and clinicopathological value of PBRM1 expression in renal cell carcinoma. Clinica Chimica Acta, Vol. 486, 2018, p. 9-17.
-
Kim J. Y., Lee S. H., Moon K. C., Kwak C., Kim H. H., Keam B., Kim T. M., Heo D. S. The impact of PBRM1 expression as a prognostic and predictive marker in metastatic renal cell carcinoma. Journal of Urology, Vol. 194, Issue 4, 2015, p. 1112-1119.
-
Piva F., Santoni M., Matrana M. R., Satti S., Giulietti M., Occhipinti G., Massari F., Cheng L., Lopez Beltran A., Scarpelli M., Principato G., Cascinu S., Montironi R. BAP1, PBRM1 and SETD2 in clear-cell renal cell carcinoma: Molecular diagnostics and possible targets for personalized therapies. Expert Review of Molecular Diagnostics, Vol. 15, Issue 9, 2015, p. 1201-1210.
-
Brugarolas J. PBRM1 and BAP1 as novel targets for renal cell carcinoma. Cancer Journal, Vol. 19, Issue 4, 2013, p. 324-332.
-
Pawłowski R., Mühl S. M., Sulser T., Krek W., Moch H., Schram P. Loss of PBRM1 expression is associated with renal cell carcinoma progression. International Journal of Cancer, Vol. 132, Issue 2, 2013, p. 27822.
-
Zhu Z., Albadawy E., Saha A., Zhang J., Harowicz M. R., Mazurowski M. A. Deep learning for identifying radiogenomic associations in breast cancer. Computers in Biology and Medicine, Vol. 109, 2019, p. 85-90.
-
Shinagare A. B., Vikram R., Jaffe C., Akin O., Kirby J., Huang E., Silverman S. G. Radiogenomics of clear cell renal cell carcinoma: preliminary findings of the cancer genome atlas-renal cell carcinoma (TCGA-RCC) imaging research group. Abdominal Imaging, Vol. 40, Issue 6, 2015, p. 1684-1692.
-
Xi Chen, Zhou Zhiguo, Hannan Raquibul, Kimberly Thomas, Pedrosa Ivan, Kapur Payal, James Brugarolas, Mou Xuanqin, Wang Jing Reliable gene mutation prediction in clear cell renal cell carcinoma through multi-classifier multi-objective radiogenomics model. Physics in Medicine and Biology, Vol. 63, Issue 21, 2018, p. 215008.
-
Kocak Burak, Emine Sebnem Durmaz, Ece Ates, Melis Baykara Ulusan Radiogenomics in clear cell renal cell carcinoma: machine learning-based high-dimensional quantitative CT texture analysis in predicting PBRM1 mutation status. American Journal of Roentgenology, Vol. 212, Issue 3, 2019, p. 55-63.
-
Eskıdere Ömer, Ali Karatutlu, Cevat Ünal Detection of Parkinson’s disease from vocal features using random subspace classifier ensemble. 12th International Conference on Electronics Computer and Computation, 2015.
-
Akin O., Elnajjar P., Heller M., Jarosz R., Erickson B. J., Kirk Filippini S. J. Radiology data from the cancer genome atlas kidney renal clear cell carcinoma [TCGA-KIRC] collection. The Cancer Imaging Archive, 2016, http://doi.org/10.7937/K9/TCIA.2016.V6PBVTDR.
-
Clark K., Vendt B., Smith K., Freymann J., Kirby J., Koppel P., Moore S., Phillips Maffitt S. D., Pringle M., Tarbox L., Prior F. The cancer imaging archive (TCIA): maintaining and operating a public information repository. Journal of Digital Imaging, Vol. 26, Issue 6, 2013, p. 1045-1057.
-
Schneider C. A., Rasband W. S., Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods, Vol. 9, Issue 7, 2012, p. 671-675.
-
Cabrera J. E. Texture Analyzer, https://imagej.nih.gov/ij/plugins/texture.html.
-
About MIPAV. Center for Information Technology, https://mipav.cit.nih.gov/.
-
Nioche C., Orlhac Boughdad Reuze Goya Outi F.-S.-S.-J., Robert C., Pellot Barakat C., Soussan M., Erique Frouin F., Buvat I. Lifex: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Research, Vol. 78, Issue 16, 2018, p. 4786-4789.
-
He Haibo, Yang Bai, Garcia Edwardo A., Li S. ADASYN: adaptive synthetic sampling approach for imbalanced learning. Journal fuer Oberflaechentechnik, Vol. 42, Issue 5, 2002, p. 56-57.
-
Yang W., Wang K., Zuo W. Neighborhood component feature selection for highdimensional data. Journal of Computers, Vol. 7, Issue 1, 2012, p. 162-168.
-
Miao D., Margolis Ca, Gao W., et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science, Vol. 359, Issue 2018, 2018, p. 801-806.
About this article
The results shown in this study are based on data made available within the by the TCGA Research Network: http://cancergenome.nih.gov/.
